If the cells you would like to access are currently listed as unavailable or
you would like information on local partners in USA, Australia, New Zealand or Japan who can support order and delivery,
please get in touch via
Contact@EBiSC.org.
BIONi010-B
K2P53, BIONi010-B
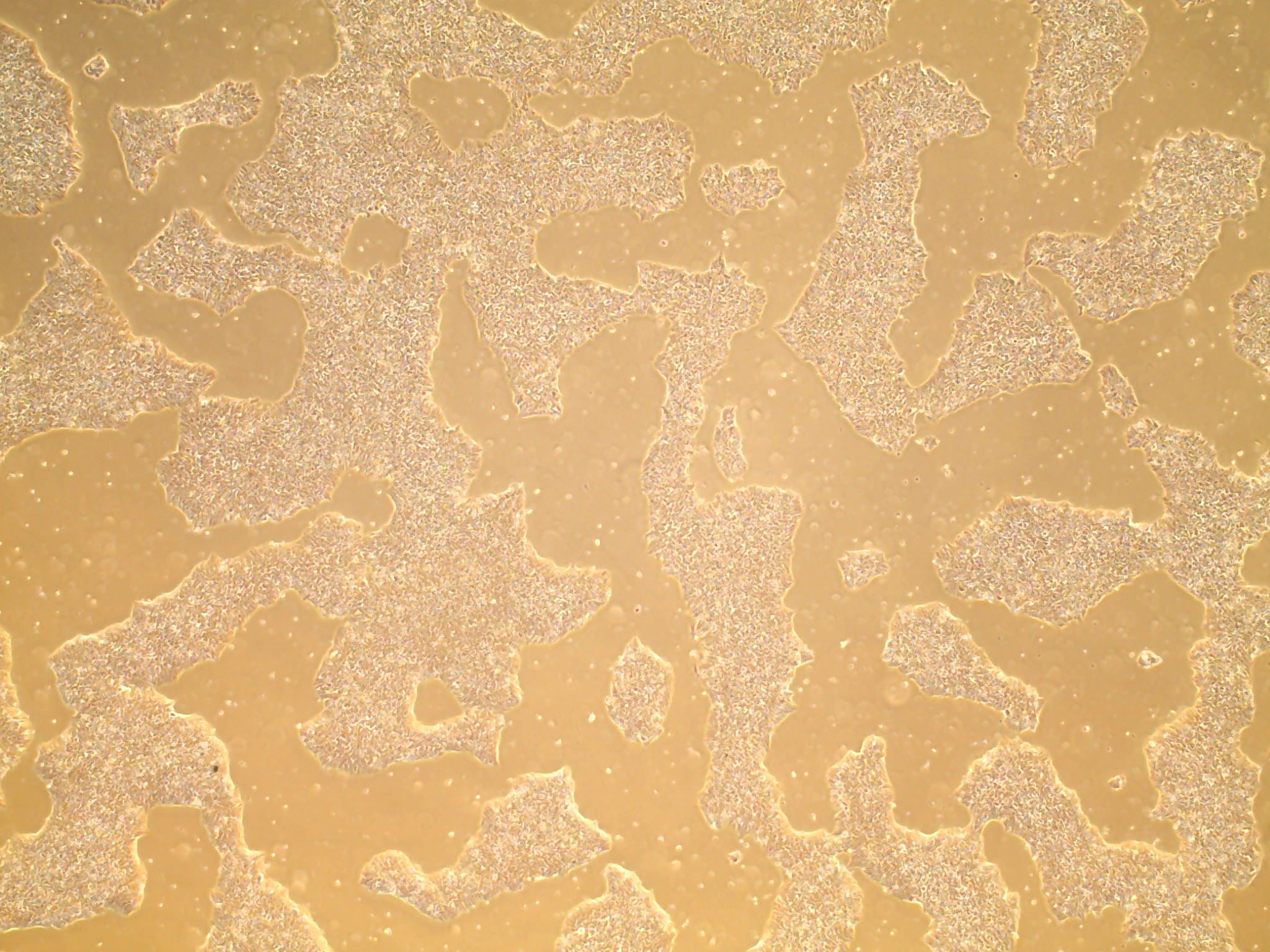
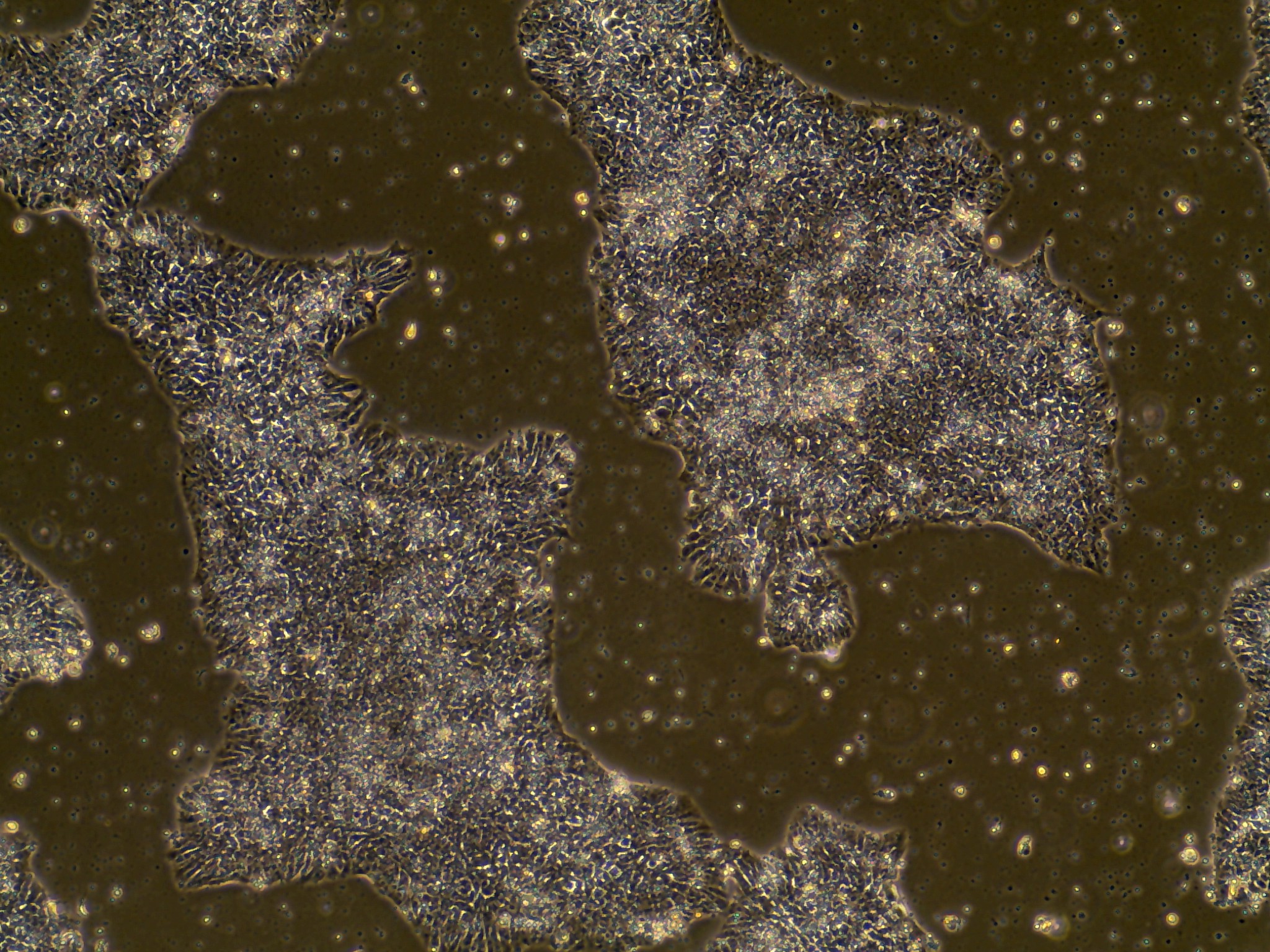
iPSC line
A CLIP contains information about a cell line including any
specific third party obligations relating to, for example,
licensing obligations or the donor consent which affect the
use of the cell line.
The EBiSC Access and Use Agreement must be completed along with an individual
Cell Line Information Pack for each line. Complete the EAUA and send to Contact@EBiSC.org
for countersignature. The EAUA must be fully signed before proceeding with your order.
A batch specific Certificate of Analysis will be available to
download once you receive your EBiSC iPSC line.
General#
Cell Line |
|
| hPSCreg name | BIONi010-B |
| Alternative name(s) |
K2P53, BIONi010-B
|
| Cell line type | Human induced pluripotent stem cell (hiPSC) |
| Similar lines |
BIONi010-A (K1P53) BIONi010-C (BIONi010-C, K3P53) BIONi010-C-71 (BIONi010-C with an APOE 3/3 genotype with an additional, homozygous christchurch mutation) BIONi010-C-54 (BIONi010-C with an APOE 4/4 genotype (with two functional alleles in contrast to BIONi010-C-4)) BIONi010-C-55 (BIONi010-C TNNI3-mCherry/TNNI1-EGFP dual reporter cl. 74) BIONi010-C-24 (BIONi010-C Dox a-syn) BIONi010-C-45 (BIONi010-C iCRE AAVS1 GBA1 LoxP EX5-6) BIONi010-C-4 (BIONi010-C ApoE E4/E4 #B44 P27) BIONi010-C-42 (BIONi010-C + iCRE AAVS1) BIONi010-C-5 (BIONi010-C CD33 E2del #N14 P26) BIONi010-C-13 (BIONi010-C + NGN2 #I7-26) BIONi010-C-2 (BIONi010-C ApoE E3/E3 #H8 P32) BIONi010-C-18 (BIONi010-C TBK1 KO) BIONi010-C-3 (BIONi010-C ApoE KO #KO30 P30) BIONi010-C-19 (BIONi010-C IKBKE KO) BIONi010-C-43 (BIONi010-C + aSNCA-wt AAVS1) BIONi010-C-15 (BIONi010-C +dox inducible NGN2-GFP) BIONi010-C-44 (BIONi010-C + aSNCA-A53T AAVS1) BIONi010-C-25 (BIONi010-C heterozygous TREM2 KO) BIONi010-C-6 (BIONi010-C ApoE E2/E2) BIONi010-C-7 (BIONi010-C Trem2 R47H) BIONi010-C-8 (BIONi010-C Trem2 T66M, #Y5-80) BIONi010-C-9 (BIONi010-C CD33 KO) BIONi010-C-49 (BIONi010-C + synapsin-m2rtTA + SNCA-wt) BIONi010-C-17 (BIONi010-C TREM2 KO) BIONi010-C-50 (BIONi010-C + synapsin-m2rtTA + SNCA-A53T) BIONi010-C-52 (BIONi010-C with an APOE 2/2 genotype (with two functional alleles in contrast to BIONi010-C-6)) BIONi010-C-70 (BIONi010-C with an APOE 2/2 genotype with an additional, homozygous christchurch mutation) BIONi010-C-53 (BIONi010-C with an APOE 3/3 genotype (with two functional alleles in contrast to BIONi010-C-2)) BIONi010-C-51 (BIONi010-C TNNI3-mCherry reporter) BIONi010-C-10 (HNF1AP291fsinsC +/- 54-5) BIONi010-C-11 (HNF1AP291fsinsC -/- 66-1) BIONi010-C-12 (HNF4ApR309C -/- 2-4) |
Provider |
|
| Depositor | Bioneer (BION) |
| Owner | Bioneer (BSC) |
| Distributors |
Bioneer (BSC)
EBiSC
|
| Derivation country | Denmark |
External Databases |
|
| hPSCreg | BIONi010-B |
| BioSamples | SAMEA3158000 |
| Cellosaurus | CVCL_1E67 |
| Wikidata | Q54796747 |
General Information |
|
| Publications | |
| This EBiSC line can be used for: |
Yes
Research use: allowed
Clinical use: no
Commercial use: no
|
Donor Information#
General Donor Information |
|
| Sex | male |
| Age of donor (at collection) | 15-19 |
| Ethnicity | Black or African-American |
Phenotype and Disease related information (Donor) |
|
| Diseases | No disease was diagnosed.
|
Other Genotyping (Donor) |
|
| Is there genome-wide genotyping or functional data available? |
No
|
Donor Relations |
|
| Other cell lines of this donor | |
External Databases (Donor) |
|
| BioSamples | SAMEA3105780 |
hIPSC Derivation#
General |
|
| Source cell line name |
CC-2511 Derived from same source line (potentially other lot and donor, see below):
|
| Source cell type |
A connective tissue cell which secretes an extracellular matrix rich in collagen and other macromolecules. Flattened and irregular in outline with branching processes; appear fusiform or spindle-shaped.; These cells may be vimentin-positive, fibronectin-positive, fsp1-positive, MMP-1-positive, collagen I-positive, collagen III-positive, and alpha-SMA-negative.
|
| Source cell origin |
Any portion of the organ that covers that body and consists of a layer of epidermis and a layer of dermis.
Synonyms
|
| Source cell type (free text) | NHDF-Ad-Der Fibroblasts |
| Source cell line lot number |
0000208364
Derived from same source line lot:
|
| Age of donor (at collection) | 15-19 |
| Source cell line vendor | LONZA |
Reprogramming method |
|
| Vector type | Non-integrating |
| Vector | Episomal |
| Genes | |
Vector free reprogramming |
|
Other |
|
| Derived under xeno-free conditions |
No |
| Derived under GMP? |
No |
| Available as clinical grade? |
No |
Culture Conditions#
Latest released batch |
|
| Culture medium | mTeSR |
| Passage method | EDTA |
| Surface coating | Matrigel / Geltrex |
| O2 concentration | 21 |
| CO2 concentration | 5 |
| Temperature | 37 |
The following are the depositor culture conditions, they do not refer to any specific batch.
| Surface coating | Matrigel/Geltrex |
| Feeder cells |
No |
| Passage method |
Enzymatically
Dispase
|
| O2 Concentration | 5 % |
| CO2 Concentration | 5 % |
| Medium |
mTeSR™ 1
|
Characterisation#
Analysis of Undifferentiated Cells
| Marker | Expressed | Immunostaining | RT-PCR | Flow Cytometry | Enzymatic Assay | Expression Profiles |
| NANOG |
Yes |
|
|
|||
| POU5F1 (OCT-4) |
Yes |
|
|
|||
| SSEA-3 |
Yes |
|
||||
| SSEA-4 |
Yes |
|
||||
| TRA 1-60 |
Yes |
|
||||
| TRA 1-81 |
Yes |
|
||||
| DNMT3B |
Yes |
|
||||
| SOX2 |
Yes |
|
Differentiation Potency
In vitro spontaneous differentiation
In vitro directed differentiation
| Marker | Expressed |
| Tuj1 |
Yes |
| NES |
Yes |
| VIMENTIN |
Yes |
| vGLUT1 |
Yes |
Microbiology / Virus Screening |
|
| HIV 1 | Negative |
| HIV 2 | Negative |
| Hepatitis B | Negative |
| Hepatitis C | Negative |
| Mycoplasma | Negative |
Sterility |
|
| Inoculation for microbiological growth | No Contaminants Detected |
| Mycoplasma | Not Detected |
| Viability | Viable post-cryopreservation |
Genotyping#
Karyotyping (Cell Line) |
|
| Has the cell line karyotype been analysed? |
Yes
|
Other Genotyping (Cell Line) |
|
| Is there genome-wide genotyping or functional data available? |
Yes
Whole genome sequencing
https://ega-archive.org/studies/EGAS00001002755 This cell line has undergone WGS using the Illumina HiSeq X platform at 30x coverage. Fastq files are stored at the European Genome Archive, users can apply for access to this data by submitting an application form to the EBiSC Data Access Committee https://ega-archive.org/dacs/EGAC00001000768 |