If the cells you would like to access are currently listed as unavailable or
you would like information on local partners in USA, Australia, New Zealand or Japan who can support order and delivery,
please get in touch via
Contact@EBiSC.org.
UKBi010-A
iLB-DS-95f-r8
iPSC line
The cell line was withdrawn.
General#
Cell Line |
|
| hPSCreg name | UKBi010-A |
| Alternative name(s) |
iLB-DS-95f-r8
|
| Cell line type | Human induced pluripotent stem cell (hiPSC) |
| Similar lines |
|
Provider |
|
| Depositor | Universitätsklinikum Bonn (UKB) |
| Owner | Institut für Rekonstruktive Neurobiologie |
| Distributors |
EBiSC
Institut für Rekonstruktive Neurobiologie
Scottish Centre for Regenerative Medicine
|
| Derivation country | Germany |
External Databases |
|
| hPSCreg | UKBi010-A |
| BioSamples | SAMEA104008300 |
| Cellosaurus | CVCL_IU29 |
| Wikidata | Q54990266 |
General Information |
|
| This EBiSC line can be used for: |
Yes
Research use: allowed
Clinical use: no
Commercial use: no
|
Donor Information#
General Donor Information |
|
| Sex | female |
| Age of donor (at collection) | 5-9 |
Phenotype and Disease related information (Donor) |
|
| Diseases | A disease was diagnosed.
|
External Databases (Donor) |
|
| BioSamples | SAMEA104010737 |
hIPSC Derivation#
General |
|
| Source cell type |
A connective tissue cell which secretes an extracellular matrix rich in collagen and other macromolecules. Flattened and irregular in outline with branching processes; appear fusiform or spindle-shaped.; These cells may be vimentin-positive, fibronectin-positive, fsp1-positive, MMP-1-positive, collagen I-positive, collagen III-positive, and alpha-SMA-negative.
|
| Source cell origin |
Any portion of the organ that covers that body and consists of a layer of epidermis and a layer of dermis.
Synonyms
|
| Age of donor (at collection) | 5-9 |
Reprogramming method |
|
| Vector type | Integrating |
| Vector | Virus (Retrovirus) |
| Genes | |
| Is the used vector excisable? |
No |
| Absence of reprogramming vector(s)? |
Unknown |
| Reprogramming vectors silenced? |
Yes |
Vector free reprogramming |
|
| Type of used vector free reprogramming factor(s) |
None
|
Other |
|
| Selection criteria for clones | Morphology |
| Derived under xeno-free conditions |
No |
| Derived under GMP? |
No |
| Available as clinical grade? |
No |
Culture Conditions#
The following are the depositor culture conditions, they do not refer to any specific batch.
| Surface coating | Matrigel/Geltrex |
| Feeder cells |
No |
| Passage method |
Enzyme-free cell dissociation
EDTA
|
| O2 Concentration | 21 % |
| CO2 Concentration | 5 % |
| Medium |
mTeSR™ 1
|
| Has Rock inhibitor (Y27632) been used at passage previously with this cell line? | No |
| Has Rock inhibitor (Y27632) been used at cryo previously with this cell line? | No |
| Has Rock inhibitor (Y27632) been used at thaw previously with this cell line? | No |
Characterisation#
Analysis of Undifferentiated Cells
| Marker | Expressed | Immunostaining | RT-PCR | Flow Cytometry | Enzymatic Assay | Expression Profiles |
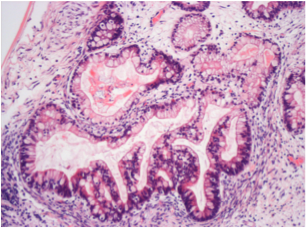
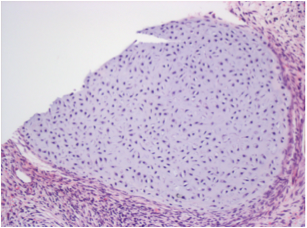
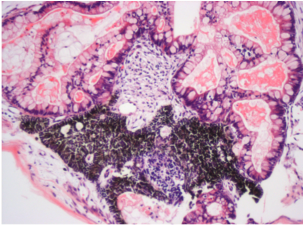
Differentiation Potency
Microbiology / Virus Screening |
|
| HIV 1 | Negative |
| HIV 2 | Negative |
| Hepatitis B | Negative |
| Hepatitis C | Negative |
| Mycoplasma | Negative |
Genotyping#
Karyotyping (Cell Line) |
|
| Has the cell line karyotype been analysed? |
No
|
Other Genotyping (Cell Line) |
|
| Is there genome-wide genotyping or functional data available? |
Yes
|