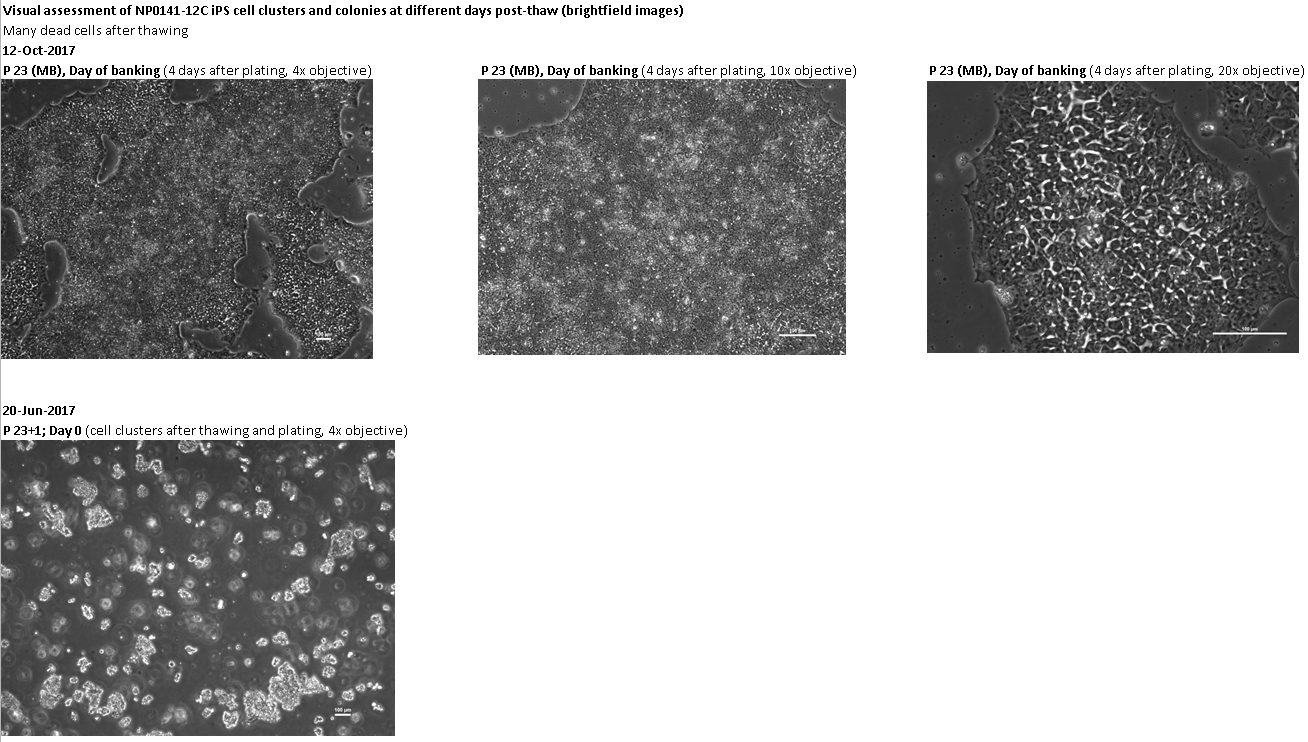
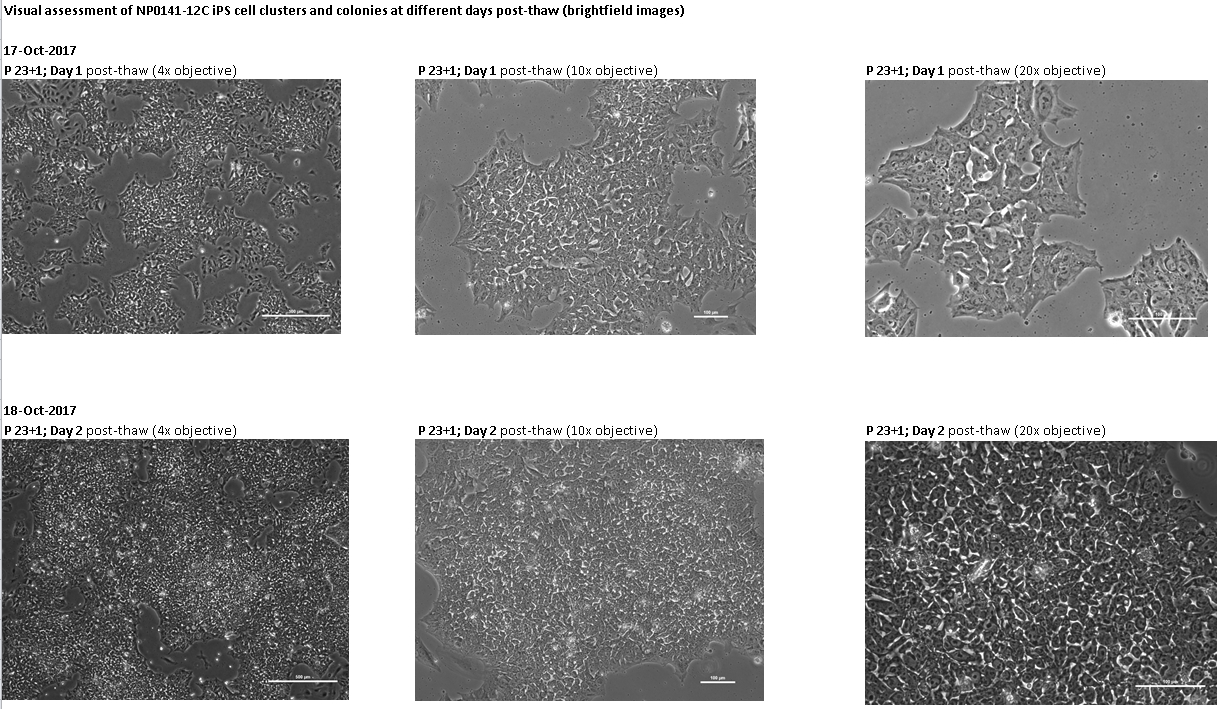
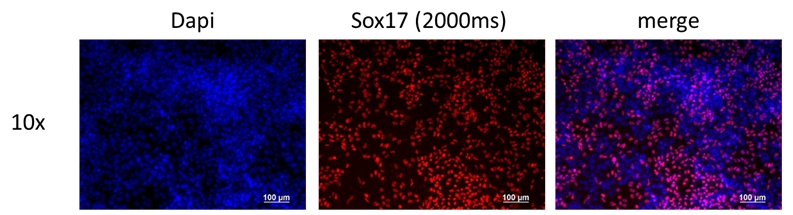
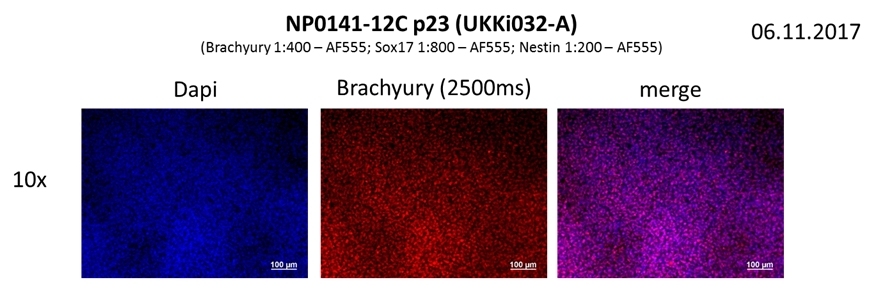
If the cells you would like to access are currently listed as unavailable or
you would like information on local partners in USA, Australia, New Zealand or Japan who can support order and delivery,
please get in touch via
Contact@EBiSC.org.
UKKi032-A
NP0141-12C
iPSC line
A CLIP contains information about a cell line including any
specific third party obligations relating to, for example,
licensing obligations or the donor consent which affect the
use of the cell line.
The EBiSC Access and Use Agreement must be completed along with an individual
Cell Line Information Pack for each line. Complete the EAUA and send to Contact@EBiSC.org
for countersignature. The EAUA must be fully signed before proceeding with your order.
To receive the Certificate of Analysis, please contact us.
A batch specific Certificate of Analysis will be available to
download once you receive your EBiSC iPSC line.
General#
Cell Line |
|
| hPSCreg name | UKKi032-A |
| Alternative name(s) |
NP0141-12C
|
| Cell line type | Human induced pluripotent stem cell (hiPSC) |
| Similar lines | |
Provider |
|
| Depositor | Klinikum der Universität zu Köln (UKK) |
| Owner | Institute for Neurophysiology, Medical Faculty |
| Distributors |
EBiSC
|
| Derivation country | Germany |
External Databases |
|
| hPSCreg | UKKi032-A |
| BioSamples | SAMEA104494417 |
| Cellosaurus | CVCL_RM77 |
| Wikidata | Q54990530 |
General Information |
|
| This EBiSC line can be used for: |
Yes
Research use: allowed
Clinical use: no
Commercial use: no
|
Donor Information#
General Donor Information |
|
| Sex | female |
| Age of donor (at collection) | 40-44 |
Phenotype and Disease related information (Donor) |
|
| Diseases | No disease was diagnosed.
|
Karyotyping (Donor) |
|
| Has the donor karyotype been analysed? |
No
|
Other Genotyping (Donor) |
|
| Is there genome-wide genotyping or functional data available? |
No
|
Donor Relations |
|
| Other cell lines of this donor | |
External Databases (Donor) |
|
| BioSamples | SAMEA104494418 |
hIPSC Derivation#
General |
|
| Source cell type |
A leukocyte with a single non-segmented nucleus in the mature form found in the circulatory pool of blood.
|
| Age of donor (at collection) | 40-44 |
| Collected in | 2016 |
| Passage number reprogrammed | 0 |
Reprogramming method |
|
| Vector type | Non-integrating |
| Vector | Sendai virus |
| Genes | |
| Is reprogramming vector detectable? |
No |
| Methods used |
PCR
|
| Files and images showing reprogramming vector expressed or silenced | |
Vector free reprogramming |
|
| Type of used vector free reprogramming factor(s) |
None
|
Other |
|
| Selection criteria for clones | morphology |
| Derived under xeno-free conditions |
Yes |
| Derived under GMP? |
No |
| Available as clinical grade? |
No |
Culture Conditions#
The following are the depositor culture conditions, they do not refer to any specific batch.
| Surface coating | Vitronectin |
| Feeder cells |
No |
| Passage method |
Enzyme-free cell dissociation
EDTA
|
| O2 Concentration | 20 % |
| CO2 Concentration | 5 % |
| Medium |
Other medium:
Base medium: StemFlex
Main protein source: Albumine Serum concentration: 0 % |
| Has Rock inhibitor (Y27632) been used at passage previously with this cell line? | No |
| Has Rock inhibitor (Y27632) been used at cryo previously with this cell line? | No |
| Has Rock inhibitor (Y27632) been used at thaw previously with this cell line? | No |
Characterisation#
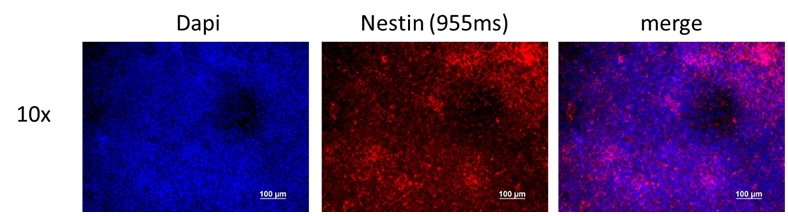
Analysis of Undifferentiated Cells
Differentiation Potency
Microbiology / Virus Screening |
|
| HIV 1 | Negative |
| HIV 2 | Negative |
| Hepatitis B | Negative |
| Hepatitis C | Negative |
| Mycoplasma | Negative |
Genotyping#
Karyotyping (Cell Line) |
|
| Has the cell line karyotype been analysed? |
Yes
No larger chromosomal aberrations to be reported; Chr12: Duplication of telomeric
Passage number: 22
|
Other Genotyping (Cell Line) |
|
| Is there genome-wide genotyping or functional data available? |
Yes
|